When a multinational specialty ingredients company set out to modernize its laboratories, it wasn’t struggling with the science—it was struggling with the plumbing. The teams were generating high‑quality fermentation, enzymology, and plant‑based nutrition research, yet the underlying data lived in too many places and too many formats. Experimental notes were captured in one electronic system, batch results were logged in another, sensitive sensory data ended up in shared drives, and strain metadata was tucked away in local databases that only a few people knew how to query. Analytical reports depended on exporting from a chromatography data system into spreadsheets, massaging peak tables and overlays by hand, and re‑keying numbers into slide decks. As sites in different regions ramped up programs in parallel, a familiar pattern emerged: duplicate experiments, inconsistent recipe transfers between bench and pilot, and QA teams losing hours preparing evidence trails that cut across multiple tools.
The company’s leadership framed the goal in plain terms: keep the core lab and enterprise systems that already work well, but add a unifying layer that standardizes data, automates capture from instruments and ERP/PLM, and enforces governance from the moment a scientist hits “start” to the moment a customer receives a certificate of analysis. The answer they chose was Scispot’s Lab OS, paired with lightweight data pipelines that could watch instrument export folders, parse files without brittle scripts, and push everything into a relational data backbone with lineage.

The transformation began with language, not code. The implementation team sat with scientists, analysts, pilot engineers, QA specialists, and program managers to agree on canonical entities and the relationships among them. A strain became more than a text field; it became a versioned object with ancestry, source, and phenotypic notes. A formulation became a governed recipe with parameters and linked materials rather than a spreadsheet tab. An experiment gained clearly defined inputs and outputs, a batch acquired a traceable life with lots and expiry, and a sensory panel captured context alongside scores. By insisting on consistent names, units, and controlled vocabularies across regions, the company eliminated the quiet chaos of near‑duplicates—sucrose versus table sugar, gram per liter versus g/L—and set a foundation that would make integrations feel routine rather than heroic.
With the shared model in place, the team moved to instrument and system ingestion. A small agent watched secure directories where the chromatography system exported runs, then parsed and normalized peak data and metadata before landing it in Scispot. Time‑series from fermentors—pH, dissolved oxygen, temperature—arrived alongside the method version, inoculation details, and media composition, ensuring that signal traces were never separated from the conditions that produced them. Item masters, lots, and expiry dates flowed in from ERP/PLM on a schedule, which meant every experiment referenced the correct material without manual lookups. None of this replaced the systems that already did their jobs; instead, Scispot served as the connective tissue, turning static files and siloed records into linked, queryable data.
Workflows followed. In R&D, scientists opened method‑guided Labsheets that looked familiar but behaved differently. The templates captured parameters consistently, calculated in context instead of in hidden spreadsheet cells, and offered structured places to record deviations and attach evidence. In the pilot plant, a formulation that had passed review became a version‑locked recipe with equipment settings and critical limits baked in, so undocumented tweaks didn’t slip between the lab and the line. QA teams approved steps with e‑signatures that left a clean trail, and method and SOP versions were tied to the records they influenced, so an auditor could see not only what happened but under which instructions it happened.
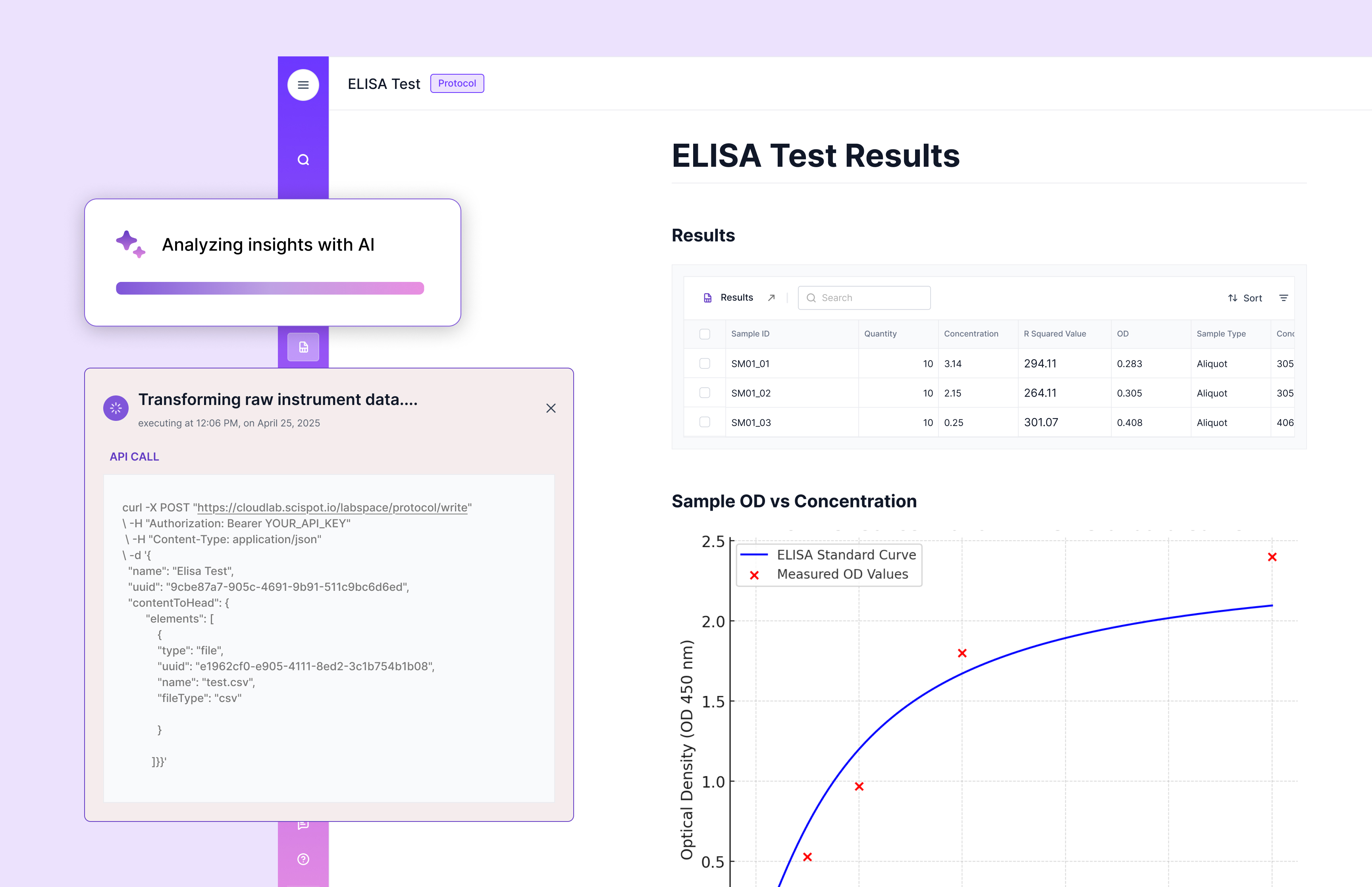
Reporting, long a tax on the analytical group’s time, changed character entirely. Rather than exporting to Excel to build overlays and area‑under‑curve summaries, analysts watched as runs appeared automatically with computed features and ready‑to‑publish figures. Reports rendered from templates that drew directly from governed data, removing the temptation to copy‑paste and the risk of accidental edits. When a study concluded, the narrative practically wrote itself: conditions, results, outliers, and links to raw and processed data were stitched together by design, not by midnight oil.

The most visible change surfaced on the dashboards that program managers and scientists came to rely on. For a fermentation series, one screen showed, in a single narrative flow, the lineage of the strain, the formulation version, the process conditions, the analytical profile, and the sensory outcome. Instead of hunting across folders and systems, stakeholders discussed the experiment, not the data locations. For operations, inventory and expiry alerts moved upstream; lots approaching expiration were flagged before they were weighed into a recipe, and low‑stock warnings popped up in time to reorder without derailing a study. For the portfolio at large, cycle‑time and right‑first‑time indicators stopped being monthly anecdotes and became continuously visible metrics that informed prioritization in real time.
Daily life changed in quiet but meaningful ways. An analytical scientist who once spent the better part of an afternoon exporting, cleaning, and formatting could now start interpreting results before lunch. A fermentation researcher no longer sent email chains to reconstruct context from a pilot run because the context traveled with the run automatically. A pilot engineer started batches knowing that the recipe was the exact version the lab intended, with deviations captured as they happened rather than as recollections afterward. A QA specialist, asked to trace everything influencing a customer batch, pulled the full story in minutes and assembled a certificate of analysis without hunting for numbers across attachments. An IT integrator, previously on call to resuscitate brittle scripts, watched standardized pipelines run, logged, and alert as expected.
The effects compounded quickly. Routine reporting and audit preparation shrank by roughly a third as manual collation disappeared and results flowed into approved templates. Go/no‑go formulation decisions grew faster; where teams once waited for end‑of‑week consolidation, many calls moved to the same day a run completed because the data was already in shape to discuss. Duplicate experiments declined as lineage and search made it obvious when a question had already been answered, even if it was answered in another region with a slightly different vocabulary. Technology transfers smoothed out; recipes crossed the bench‑to‑pilot boundary with their context intact, so right‑first‑time rates improved without requiring heroics on the floor. Compliance confidence rose because approvals, SOP versions, and audit trails were embedded into normal work, not bolted on later.
None of this required the company to abandon systems that already worked. The chromatography data system remained the source of truth for acquisition and integration. The ERP and PLM kept governing financial and product structures. Scispot filled the gaps between those systems, specialized instruments, and human workflows, emphasizing harmonization over replacement. That choice kept risk low, minimized retraining, and ensured scientists could focus on the science rather than adapting to a brand‑new stack.
The architecture behind the scenes was straightforward by design. Scispot’s Lab OS provided the data model, permissions, and approvals, while the data pipelines performed the unglamorous but crucial tasks of watching folders, parsing outputs, and validating files. The platform stored linked records rather than isolated files, which meant that an experiment, a batch, or a sensory panel was always tied to who did the work, when they did it, which method and SOP version they used, and which materials they consumed. Dashboards read from the same backbone, so visualizations didn’t drift from the underlying truth, and exporting a report became a repeatable, governed act rather than a bespoke exercise.
Change management mattered as much as technology. The company rolled out in short, targeted sprints, each one solving a visible pain point using real data rather than demo sets. Analytical reporting went first because it offered immediate time savings and a morale boost. Fermentation lineage followed because it anchored collaboration between upstream R&D and downstream pilot teams. Inventory and expiry controls came next because they paid dividends in fewer surprises. The team avoided the trap of perfecting every edge case in advance; instead, they designed for auditability from day one, accepted that templates would evolve, and treated vocabulary as a managed asset rather than a background nuisance.
Looking back, a few lessons stand out. Standardizing the nouns—strain, formulation, experiment, batch, lot, and panel—unlock more value than any single integration because it ensures every system and every person is speaking the same language. Automating the middle steps—the quiet movements of files and numbers between tools—pays back faster than glamorous analytics because it reclaims hours that teams can spend interpreting instead of assembling. Building governance into the flow of work—e‑signatures when decisions are made, versioning when methods change, audit logs that write themselves—turns compliance from a scramble into a non‑event. Keeping proven systems in place reduces risk and accelerates adoption; the thesis is not to replace what works, but to connect what doesn’t talk. And measuring decision latency—the time from “run complete” to “action taken”—keeps the program honest about whether the transformation is helping where it matters.
With a clean, linked history of strain, formulation, process, analytical, and sensory data, the company is now exploring models that predict fermentation outcomes from input parameters and hint at sensory results from analytical fingerprints. Because the data is standardized and traceable, those projects start on firm footing rather than on one‑off exports. The same backbone is informing supplier analytics at the lot level and enabling simple closed‑loop experiments, where promising results trigger follow‑ups automatically rather than waiting for a monthly review.
This story does not hinge on a single feature. It is about building a laboratory nervous system that is humble in its parts and ambitious in its effect. By unifying the day’s work into governed, searchable, and sharable records—without tearing out the tools scientists rely on—Scispot helped a global ingredients innovator cut busywork, sharpen decisions, improve tech transfer, and step confidently into AI‑ready R&D. The science was always strong. Now the data supports it with the same clarity and speed.